github.com/jaredleekatzman/DeepSurv
jaredleekatzman/DeepSurv
DeepSurv is a deep learning approach to survival analysis. - jaredleekatzman/DeepSurv
github.com
Medical practitioners use survival models to explore and understand the relationships between patients' covariates (e.g. clinical and genetic features) and the effectiveness of various treatment options. Standard survival models like the linear Cox proportional hazards model require extensive feature engineering or prior medical knowledge to model treatment interaction at an individual level. While nonlinear survival methods, such as neural networks and survival forests, can inherently model these high-level interaction terms, they have yet to be shown as effective treatment recommender systems. We introduce DeepSurv, a Cox proportional hazards deep neural network and state-of-the-art survival method for modeling interactions between a patient's covariates and treatment effectiveness in order to provide personalized treatment recommendations. We perform a number of experiments training DeepSurv on simulated and real survival data. We demonstrate that DeepSurv performs as well as or better than other state-of-the-art survival models and validate that DeepSurv successfully models increasingly complex relationships between a patient's covariates and their risk of failure. We then show how DeepSurv models the relationship between a patient's features and effectiveness of different treatment options to show how DeepSurv can be used to provide individual treatment recommendations. Finally, we train DeepSurv on real clinical studies to demonstrate how it's personalized treatment recommendations would increase the survival time of a set of patients. The predictive and modeling capabilities of DeepSurv will enable medical researchers to use deep neural networks as a tool in their exploration, understanding, and prediction of the effects of a patient's characteristics on their risk of failure.
DeepSurv
DeepSurv implements a deep learning generalization of the Cox proportional hazards model using Theano and Lasagne.
DeepSurv has an advantage over traditional Cox regression because it does not require an a priori selection of covariates, but learns them adaptively.
DeepSurv can be used in numerous survival analysis applications. One medical application is provided: recommend_treatment, which provides treatment recommendations for a set of patient observations.
For more details, see full paper DeepSurv: Personalized Treatment Recommender System Using A Cox Proportional Hazards Deep Neural Network.
For an updated implementation of the Cox loss function in PyTorch, please see Feature Selection using Stochastic Gates (STG) by Yamada et al..
Installation :
From source
Download a local copy of DeepSurv and install from the directory:
git clone https://github.com/jaredleekatzman/DeepSurv.git
cd DeepSurv
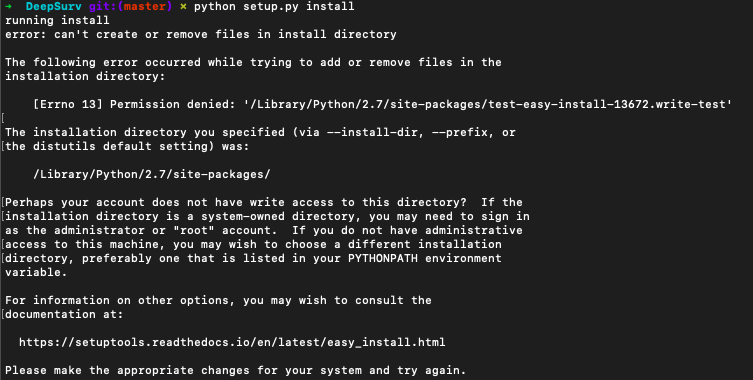
python setup.py install

성공하지 못했다.
sudo python setup.py install

성공
test_deepsurv.py

test_deepsurv를 먼저 실행시켜보았다.
test_deepsurv는 test 파일 안에 있다.
test_deepsurv.py 코드에 syntax error가 있었다.
if문의 끝에 : 콜론 붙여주어야 한다.
Requirment
| python 3.5 // 본인은 python 3.8 이용 |
| h5py>=2.7.0 |
| lifelines>=0.9.4 |
| logger==1.4 |
| Optunity>=1.1.1 |
| tensorboard_logger>=0.0.3 |
| lasagne==0.2.dev1 |
| theano>=0.8.2 |
sudo pip3 install h5py
sudo pip3 install lifelines
sudo pip3 install Optunity
sudo pip3 install tensorboard_logger
sudo pip3 install logger==1.4
sudo pip3 install lasagne
sudo pip3 install --upgrade https://github.com/Lasagne/Lasagne/archive/master.zip모든 라이브러리를 설치해야 test.py가 작동한다.
lasagne의 경우 0.1 버전으로 설치가 되므로 github에 있는 링크를 통해 0.2.dev1 로 업그레이드 해주어야 한다.
$ pip install -r requirements.txt이렇게만 쳐도 된다고 한다.
Dependencies
Theano, Lasagne (bleeding edge version), lifelines, matplotlib (for visualization), tensorboard_logger, and all of their respective dependencies.
lifelines 란?
lifelines는 Kaplan-Meier 그래프 및 logrank 함수를 제공하는 생존분석 특화 모듈이다. pip을 이용하여 손쉽게 설치할 수 있으며, 자세한 내용은 홈페이지를 참고한다.
출처 : blog.naver.com/PostView.nhn?blogId=cjh226&logNo=221266296236&parentCategoryNo=&categoryNo=17&viewDate=&isShowPopularPosts=false&from=postView
Kaplan-Meier curve는 두 환자군의 생존율을 비교분석하기 위한 생존분석(survival analysis) 기법이다. 주로 암환자의 예후(prognosis)를 비교하는데 사용되며, '특정 약물에 대한 투여군vs비투여군' 또는 '특정 유전자에 대한 과발현군vs정상군' 등에 활용된다.
Logrank test는 '두 환자군의 생존 확률 분포(survival distribution)이 동일하다'는 귀무가설(null hypothesis)를 검정한다. Kaplan-Meier 생존곡선을 나타낼 때, 두 군의 차이에 대한 유의성(P-value)도 같이 나타내도록 한다.
Matplotlib 이란?
matplotlib은 다양한 데이터를 많은 방법으로 도식화 할 수 있도록 하는 파이썬 라이브러리로써, 우리는 matplotlib의 pyplot을 이용하게 됩니다.
이는 mathworks에서 개발한 매트랩(MATLAB)과 비슷한 형태를 가지고 있습니다.
matplotlib을 이용하면 우리가 이전에 알아본 numpy나 pandas에서 사용되는 자료구조를 쉽게 시각화 할 수 있습니다.
출처: https://doorbw.tistory.com/173 [Tigercow.Door]
Running the tests
pytest를 통해 test.py 를 진행 해보았다.
먼저 pytest 라이브러리를 설치한다.
sudo pip3 install pytestpytest

앞에서 if문에 syntex error가 발생하였기 때문에 수정해준다.
test_train()에서 if문 " : " 추가
def test_train(self):
if sys.version_info.major == 2:
pytest 성공
pytest test_deepsurv.py
deep_surv.py


➜ deepsurv git:(master) ✗ python3 -m deep_surv.py
Traceback (most recent call last):
File "/Library/Developer/CommandLineTools/Library/Frameworks/Python3.framework/Versions/3.8/lib/python3.8/runpy.py", line 184, in _run_module_as_main
mod_name, mod_spec, code = _get_module_details(mod_name, _Error)
File "/Library/Developer/CommandLineTools/Library/Frameworks/Python3.framework/Versions/3.8/lib/python3.8/runpy.py", line 110, in _get_module_details
__import__(pkg_name)
File "/Users/seon-uchan/Desktop/Intern/practice/DeepSurv/deepsurv/deep_surv.py", line 14, in <module>
from .deepsurv_logger import DeepSurvLogger
ImportError: attempted relative import with no known parent packagepath 오류
from .deepsurv_logger import DeepSurvLogger↓↓↓↓↓↓↓↓↓↓↓↓↓↓↓↓↓↓↓↓↓↓↓↓↓↓↓↓↓↓↓↓↓↓↓↓↓↓↓↓↓↓↓↓↓↓↓↓↓↓↓↓↓↓↓↓↓↓↓↓↓↓
from deepsurv_logger import DeepSurvLogger
python 3.8에서는 path에 대한 문법이 살짝 다르다.
위와 같이 deepsurv_py를 수정해준다.
Docker로 Installation 할 경우
Running Experiments
Experiments are run using Docker containers built off of the floydhub deep learning Docker images. DeepSurv can be run on either the CPU or the GPU with nvidia-docker.
All experiments are in the DeepSurv/experiments/ directory.
To run an experiment, define the experiment name as an environmental variable EXPRIMENTand run the docker-compose file. Further details are in the DeepSurv/experiments/ directory.
floydhub : 제한적 무료 딥러닝 클라우드 플랫폼
(이메일로만 계정 확인을 하는 점을 생각하면, 여러 ID로 돌릴수가 있다)
DeepSurv Experiments
Experiments are run using Docker containers built off of the floydhub deep learning Docker images.
Requirements
Download and install Docker. If you plan to use FloydHub's GPU tag, install nvidia-docker.
nvidia-docker로 개발환경 한방에 세팅하기
GPU를 사용하는 머신러닝 환경을 구축하기 위해서는 virtualenv, anaconda 등 파이썬의 가상환경을 통한 다양한 방법이 제시되는데 이와 같은 환경에는 문제가 하나 있다. 바로 다양한 버전의 CUDA를 사
jybaek.tistory.com
일반 Docker를 사용할 경우 GPU에 제한이 있다. 따라서 그래픽 요소가 많이 사용되는 프로그램이나 딥러닝 같은 연산이 필요한 도커를 쓰는 경우 GPU를 이어주는 Nvidia-Docker 툴을 설치, 사용해야 한다.
Experiments
To run one of the experiments from the paper, use the following command from this directory:
EXPERIMENT=${EXPERIMENT_ID} docker-compose up --build
The following experiments are provided:
| Experiment | Experiment ID |
| Simulated Linear Data | linear |
| Simulated Nonlinear (Gaussian) Data | gaussian |
| Worchester Heart Attack Study (WHAS) | whas |
| Study to Understand Prognoses Preferences Outcomes and Risks of Treatment (SUPPORT) | support |
| Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) | metabric |
| Simulated Treatment Data | treatment |
| Rotterdam & German Breast Cancer Study Group (GBSG) | gbsg |
The hyper-parameters for each experiment are in this repo's directory:
DeepSurv/experiments/deepsurv/models/
For more details on each experiment, reference DeepSurv: Personalized Treatment Recommender System Using A Cox Proportional Hazards Deep Neural Network.
Using GPU
If you have nvidia-docker installed, you can run DeepSurv experiments using your GPU. To do so change the tag in the first line of your experiment's docker file.
For example, to run the simulated linear data experiment with the GPU change the first line the file
./deepsurv/Docker.linear to:
FROM floydhub/dl-docker:gpuTraining a Network
Training DeepSurv can be done in a few lines. First, all you need to do is prepare the datasets to have the following keys:
{
'x': (n,d) observations (dtype = float32),
't': (n) event times (dtype = float32),
'e': (n) event indicators (dtype = int32)
}
Then prepare a dictionary of hyper-parameters. And it takes only two lines to train a network:
network = deepsurv.DeepSurv(**hyperparams)
log = network.train(train_data, valid_data, n_epochs=500)
You can then evaluate its success on testing data:
network.get_concordance_index(**test_data)
>> 0.62269622730138632
If you have matplotlib installed, you can visualize the training and validation curves after training the network:
deepsurv.plot_log(log)
'📌 Paper > Deepsurv' 카테고리의 다른 글
| Deepsurv - method 및 관련 개념 (0) | 2021.01.18 |
|---|---|
| 생존 함수, Cox hazard model (0) | 2021.01.11 |
| Deepsurv 논문 읽기 (0) | 2021.01.11 |
| Tensorflow - Theano - Torch - Keras - Lasagne (0) | 2021.01.04 |
| Cox Proportional Hazards (0) | 2021.01.04 |